Functionality for k-means clustering ('kmeans') objects
Source:R/methods-stats-kmeans.r
methods-kmeans.RdThese methods extract data from, and attribute new data to,
objects of class "kmeans" as returned by stats::kmeans().
Usage
# S3 method for class 'kmeans'
as_tbl_ord(x)
# S3 method for class 'kmeans'
recover_rows(x)
# S3 method for class 'kmeans'
recover_cols(x)
# S3 method for class 'kmeans'
recover_coord(x)
# S3 method for class 'kmeans'
recover_aug_rows(x)
# S3 method for class 'kmeans'
recover_aug_cols(x)
# S3 method for class 'kmeans'
recover_aug_coord(x)Value
The recovery generics recover_*() return core model components, distribution of inertia,
supplementary elements, and intrinsic metadata; but they require methods for each model class to
tell them what these components are.
The generic as_tbl_ord() returns its input wrapped in the 'tbl_ord'
class. Its methods determine what model classes it is allowed to wrap. It
then provides 'tbl_ord' methods with access to the recoverers and hence to
the model components.
See also
Other methods for idiosyncratic techniques:
methods-lm
Other models from the stats package:
methods-cancor,
methods-cmds,
methods-factanal,
methods-lm,
methods-prcomp,
methods-princomp
Examples
# data frame of Anderson iris species measurements
class(iris)
#> [1] "data.frame"
head(iris)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3.0 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5.0 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosa
# compute 3-means clustering on scaled iris measurements
set.seed(5601L)
iris %>%
subset(select = -Species) %>%
scale() %>%
kmeans(centers = 3) %>%
print() -> iris_km
#> K-means clustering with 3 clusters of sizes 50, 53, 47
#>
#> Cluster means:
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 1 -1.01119138 0.85041372 -1.3006301 -1.2507035
#> 2 -0.05005221 -0.88042696 0.3465767 0.2805873
#> 3 1.13217737 0.08812645 0.9928284 1.0141287
#>
#> Clustering vector:
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
#> 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40
#> 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60
#> 1 1 1 1 1 1 1 1 1 1 3 3 3 2 2 2 3 2 2 2
#> 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80
#> 2 2 2 2 2 3 2 2 2 2 3 2 2 2 2 3 3 3 2 2
#> 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100
#> 2 2 2 2 2 3 3 2 2 2 2 2 2 2 2 2 2 2 2 2
#> 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120
#> 3 2 3 3 3 3 2 3 3 3 3 3 3 2 2 3 3 3 3 2
#> 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140
#> 3 2 3 2 3 3 2 3 3 3 3 3 3 2 2 3 3 3 2 3
#> 141 142 143 144 145 146 147 148 149 150
#> 3 3 2 3 3 3 2 3 3 2
#>
#> Within cluster sum of squares by cluster:
#> [1] 47.35062 44.08754 47.45019
#> (between_SS / total_SS = 76.7 %)
#>
#> Available components:
#>
#> [1] "cluster" "centers" "totss" "withinss" "tot.withinss"
#> [6] "betweenss" "size" "iter" "ifault"
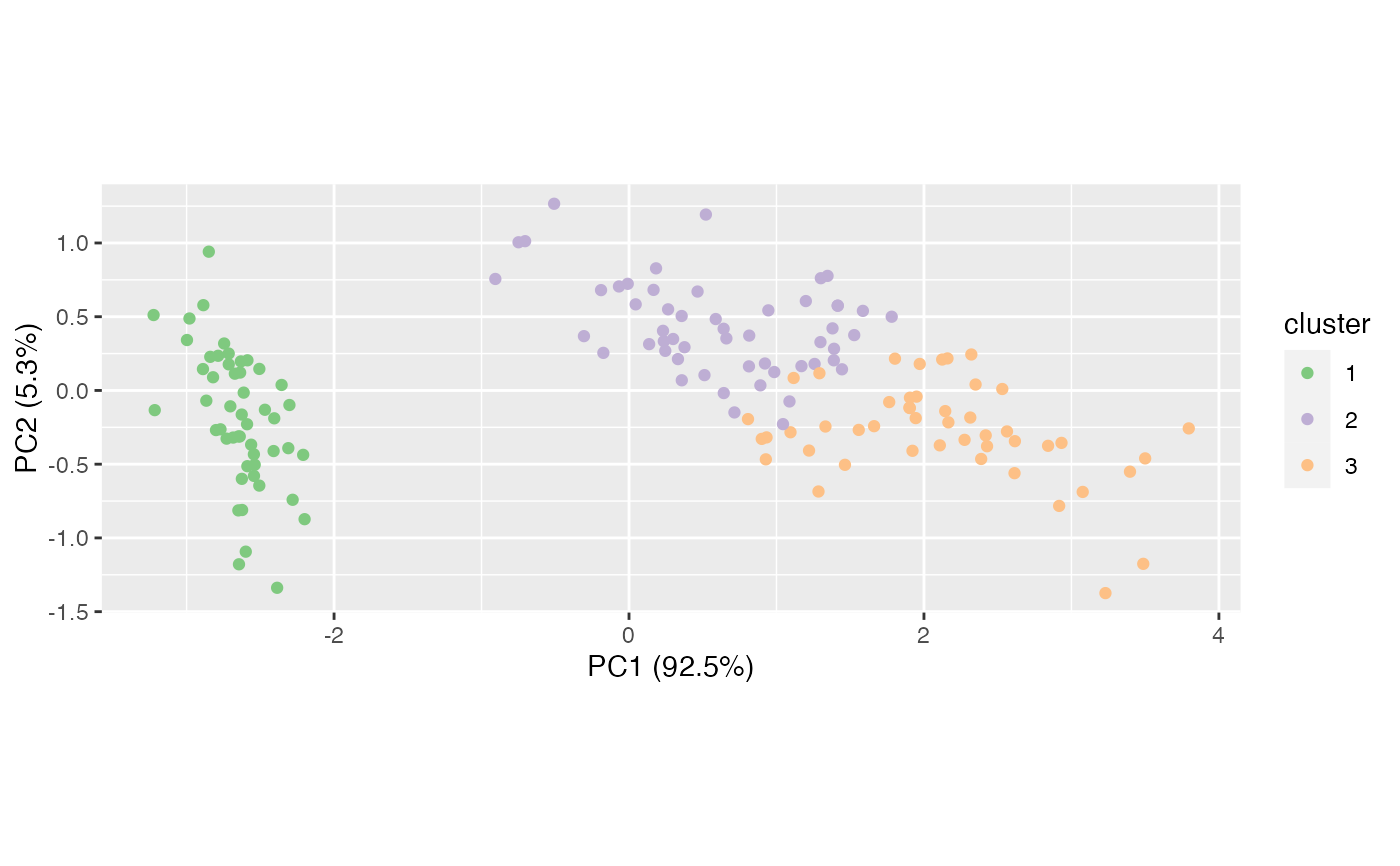
# visualize clusters using PCA
iris %>%
subset(select = -Species) %>%
prcomp() %>%
as_tbl_ord() %>%
mutate_rows(cluster = iris_km$cluster) %>%
ggbiplot() +
geom_rows_point(aes(color = factor(as.character(as.integer(cluster)),
levels = as.character(seq(3L))))) +
scale_color_brewer(type = "qual", name = "cluster")
 # wrap as a 'tbl_ord' object
(iris_km_ord <- as_tbl_ord(iris_km))
#> # A tbl_ord of class 'kmeans': (150 x 3) x (4 x 3)'
#> # 3 coordinates: 1, 2, 3
#> #
#> # Rows: [ 150 x 3 | 0 ]
#> `1` `2` `3` |
#> |
#> 1 1 0 0 |
#> 2 1 0 0 |
#> 3 1 0 0 |
#> 4 1 0 0 |
#> 5 1 0 0 |
#> # ℹ 145 more rows |
#>
#> #
#> # Columns: [ 4 x 3 | 0 ]
#> `1` `2` `3` |
#> |
#> 1 -1.01 -0.0501 1.13 |
#> 2 0.850 -0.880 0.0881 |
#> 3 -1.30 0.347 0.993 |
#> 4 -1.25 0.281 1.01 |
# augment everything with names, observations with cluster assignment
(iris_km_ord <- augment_ord(iris_km_ord))
#> # A tbl_ord of class 'kmeans': (150 x 3) x (4 x 3)'
#> # 3 coordinates: 1, 2, 3
#> #
#> # Rows: [ 150 x 3 | 2 ]
#> `1` `2` `3` | name cluster
#> | <chr> <fct>
#> 1 1 0 0 | 1 1 1
#> 2 1 0 0 | 2 2 1
#> 3 1 0 0 | 3 3 1
#> 4 1 0 0 | 4 4 1
#> 5 1 0 0 | 5 5 1
#> # ℹ 145 more rows | # ℹ 145 more rows
#>
#> #
#> # Columns: [ 4 x 3 | 1 ]
#> `1` `2` `3` | name
#> | <chr>
#> 1 -1.01 -0.0501 1.13 | 1 Sepal.Length
#> 2 0.850 -0.880 0.0881 | 2 Sepal.Width
#> 3 -1.30 0.347 0.993 | 3 Petal.Length
#> 4 -1.25 0.281 1.01 | 4 Petal.Width
# summarize clusters with standard deviation
iris_km_ord %>%
tidy() %>%
transform(sdev = sqrt(withinss / size))
#> name size withinss inertia prop_var quality sdev
#> 1 1 50 47.35062 NA NA NA 0.9731456
#> 2 2 53 44.08754 NA NA NA 0.9120529
#> 3 3 47 47.45019 NA NA NA 1.0047779
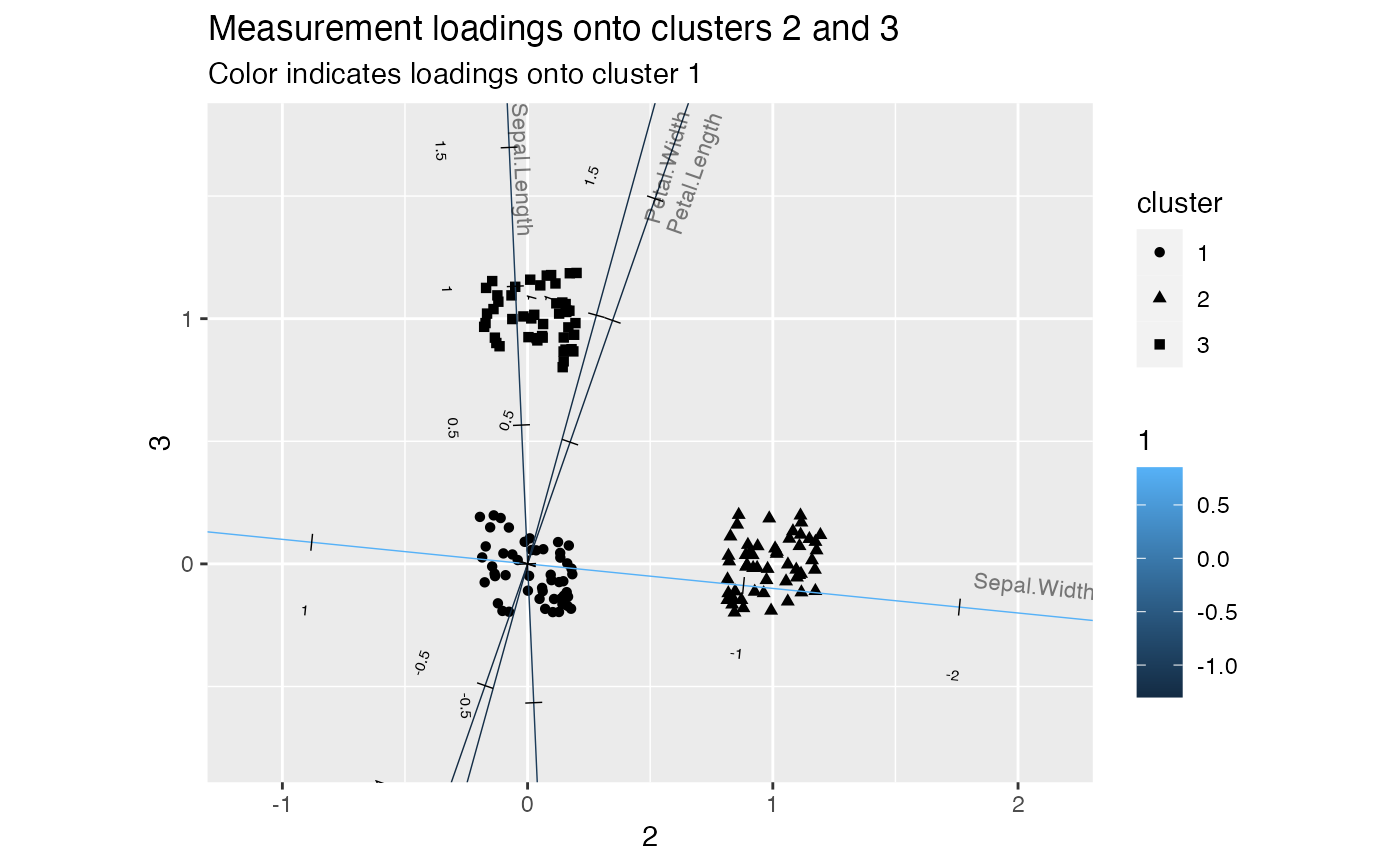
# discriminate between clusters 2 and 3
iris_km_ord %>%
ggbiplot(aes(x = `2`, y = `3`), color = factor(.cluster)) +
geom_jitter(stat = "rows", aes(shape = cluster), width = .2, height = .2) +
geom_cols_axis(aes(color = `1`, label = name),
text.size = 2, text_dodge = .1,
size = 3, label.alpha = .5) +
scale_x_continuous(expand = expansion(mult = .8)) +
scale_y_continuous(expand = expansion(mult = .5)) +
ggtitle(
"Measurement loadings onto clusters 2 and 3",
"Color indicates loadings onto cluster 1"
)
# wrap as a 'tbl_ord' object
(iris_km_ord <- as_tbl_ord(iris_km))
#> # A tbl_ord of class 'kmeans': (150 x 3) x (4 x 3)'
#> # 3 coordinates: 1, 2, 3
#> #
#> # Rows: [ 150 x 3 | 0 ]
#> `1` `2` `3` |
#> |
#> 1 1 0 0 |
#> 2 1 0 0 |
#> 3 1 0 0 |
#> 4 1 0 0 |
#> 5 1 0 0 |
#> # ℹ 145 more rows |
#>
#> #
#> # Columns: [ 4 x 3 | 0 ]
#> `1` `2` `3` |
#> |
#> 1 -1.01 -0.0501 1.13 |
#> 2 0.850 -0.880 0.0881 |
#> 3 -1.30 0.347 0.993 |
#> 4 -1.25 0.281 1.01 |
# augment everything with names, observations with cluster assignment
(iris_km_ord <- augment_ord(iris_km_ord))
#> # A tbl_ord of class 'kmeans': (150 x 3) x (4 x 3)'
#> # 3 coordinates: 1, 2, 3
#> #
#> # Rows: [ 150 x 3 | 2 ]
#> `1` `2` `3` | name cluster
#> | <chr> <fct>
#> 1 1 0 0 | 1 1 1
#> 2 1 0 0 | 2 2 1
#> 3 1 0 0 | 3 3 1
#> 4 1 0 0 | 4 4 1
#> 5 1 0 0 | 5 5 1
#> # ℹ 145 more rows | # ℹ 145 more rows
#>
#> #
#> # Columns: [ 4 x 3 | 1 ]
#> `1` `2` `3` | name
#> | <chr>
#> 1 -1.01 -0.0501 1.13 | 1 Sepal.Length
#> 2 0.850 -0.880 0.0881 | 2 Sepal.Width
#> 3 -1.30 0.347 0.993 | 3 Petal.Length
#> 4 -1.25 0.281 1.01 | 4 Petal.Width
# summarize clusters with standard deviation
iris_km_ord %>%
tidy() %>%
transform(sdev = sqrt(withinss / size))
#> name size withinss inertia prop_var quality sdev
#> 1 1 50 47.35062 NA NA NA 0.9731456
#> 2 2 53 44.08754 NA NA NA 0.9120529
#> 3 3 47 47.45019 NA NA NA 1.0047779
# discriminate between clusters 2 and 3
iris_km_ord %>%
ggbiplot(aes(x = `2`, y = `3`), color = factor(.cluster)) +
geom_jitter(stat = "rows", aes(shape = cluster), width = .2, height = .2) +
geom_cols_axis(aes(color = `1`, label = name),
text.size = 2, text_dodge = .1,
size = 3, label.alpha = .5) +
scale_x_continuous(expand = expansion(mult = .8)) +
scale_y_continuous(expand = expansion(mult = .5)) +
ggtitle(
"Measurement loadings onto clusters 2 and 3",
"Color indicates loadings onto cluster 1"
)