Compute normal confidence ellipses for ordination factors
stat-biplot-ellipse.RdThese ordination stats are adapted from
ggplot2::stat_ellipse().
stat_rows_ellipse( mapping = NULL, data = NULL, geom = "path", position = "identity", show.legend = NA, inherit.aes = TRUE, ..., type = "t", level = 0.95, segments = 51 ) stat_cols_ellipse( mapping = NULL, data = NULL, geom = "path", position = "identity", show.legend = NA, inherit.aes = TRUE, ..., type = "t", level = 0.95, segments = 51 )
Arguments
| mapping | Set of aesthetic mappings created by |
|---|---|
| data | The data to be displayed in this layer. There are three options: If A A |
| geom | The geometric object to use display the data |
| position | Position adjustment, either as a string, or the result of a call to a position adjustment function. |
| show.legend | logical. Should this layer be included in the legends?
|
| inherit.aes | If |
| ... | Additional arguments passed to |
| type | The type of ellipse.
The default |
| level | The level at which to draw an ellipse,
or, if |
| segments | The number of segments to be used in drawing the ellipse. |
Format
An object of class StatRowsEllipse (inherits from StatEllipse, Stat, ggproto, gg) of length 2.
An object of class StatColsEllipse (inherits from StatEllipse, Stat, ggproto, gg) of length 2.
Biplot layers
ggbiplot() uses ggplot2::fortify() internally to produce a single data
frame with a .matrix column distinguishing the subjects ("rows") and
variables ("cols"). The stat layers stat_rows() and stat_cols() simply
filter the data frame to one of these two.
The geom layers geom_rows_*() and geom_cols_*() call the corresponding
stat in order to render plot elements for the corresponding factor matrix.
geom_dims_*() selects a default matrix based on common practice, e.g.
points for rows and arrows for columns.
Examples
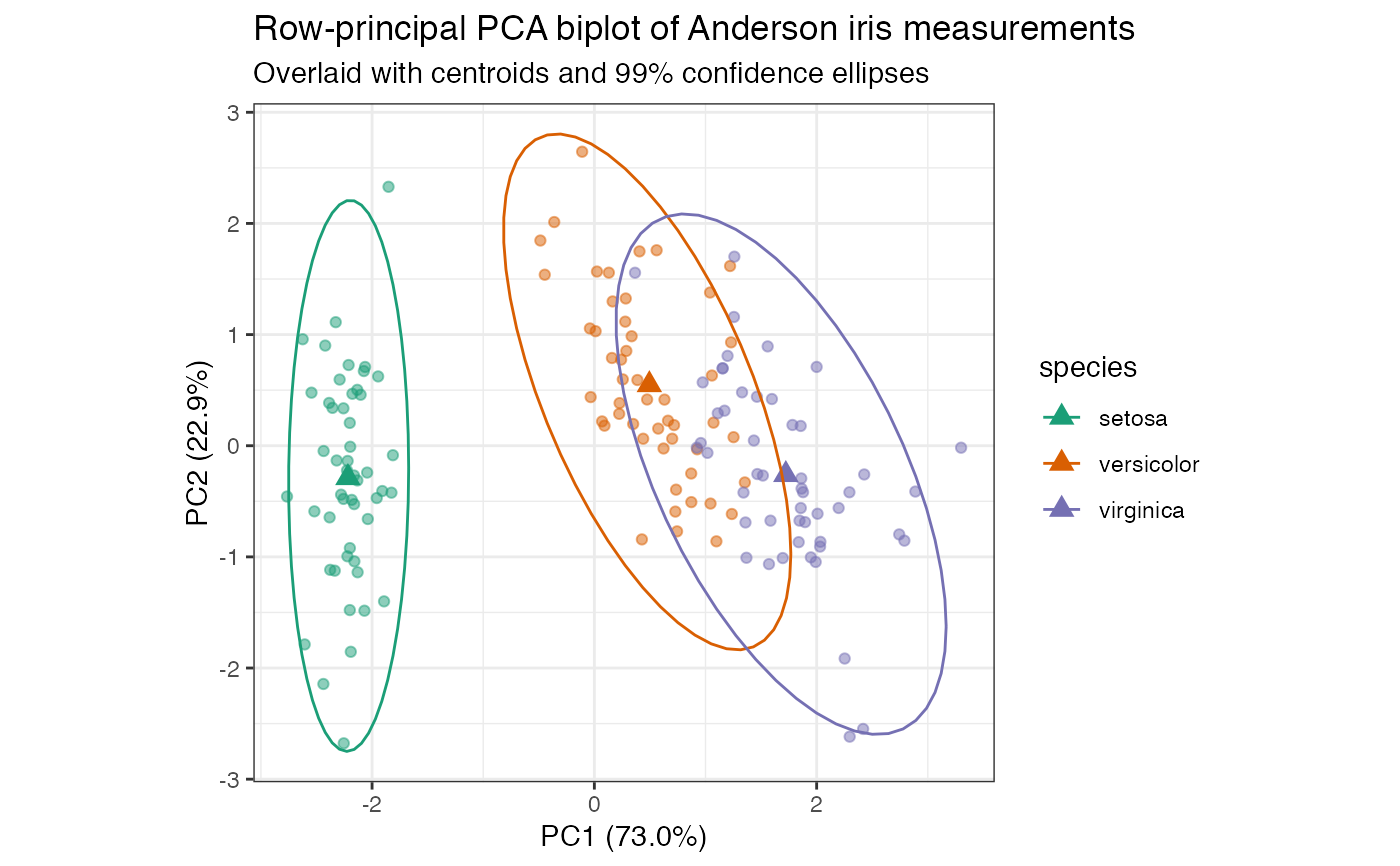
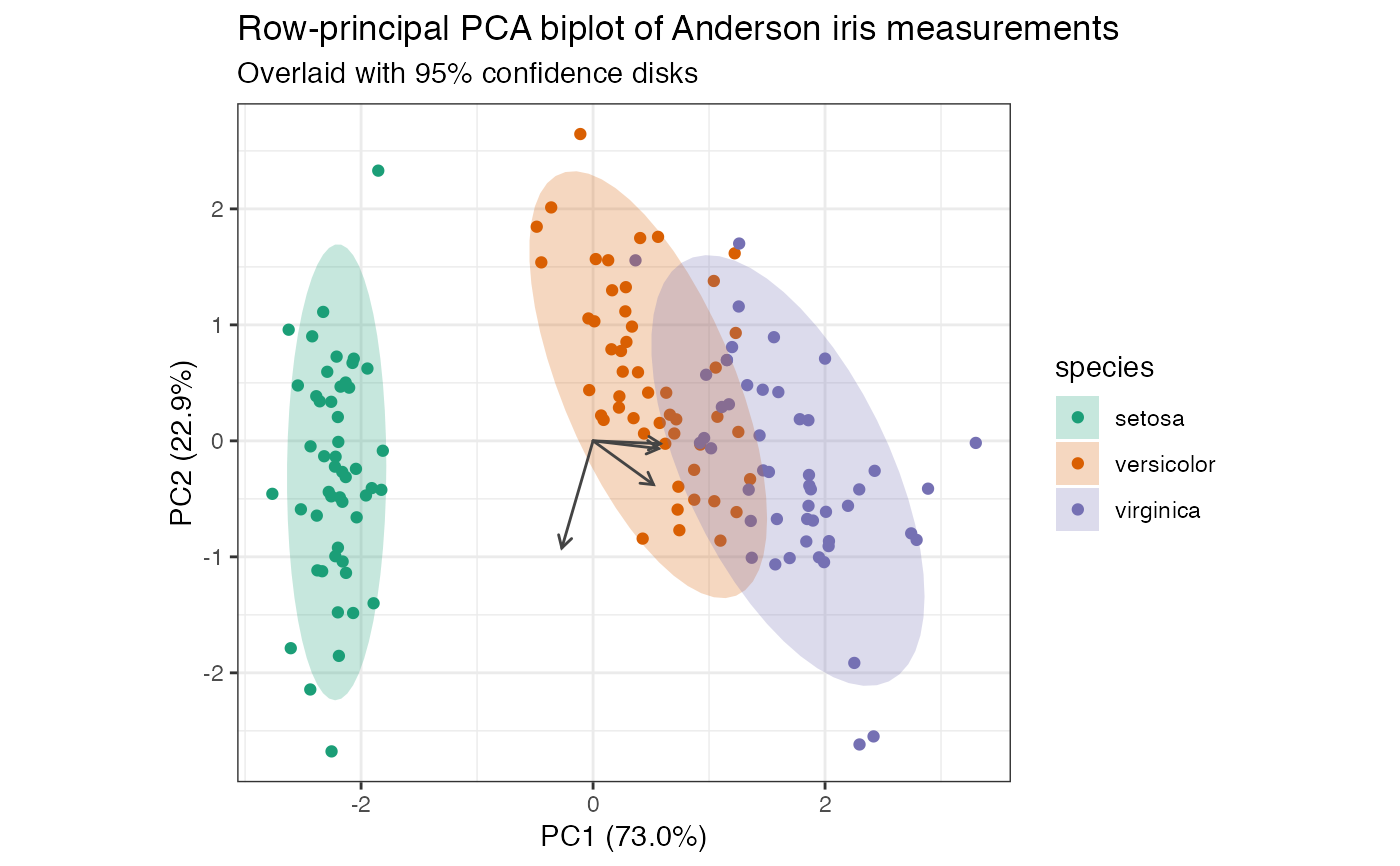
# compute row-principal components of scaled iris measurements iris[, -5] %>% prcomp(scale = TRUE) %>% as_tbl_ord() %>% mutate_rows(species = iris$Species) %>% print() -> iris_pca#> # A tbl_ord of class 'prcomp': (150 x 4) x (4 x 4)' #> # 4 coordinates: PC1, PC2, ..., PC4 #> # #> # Rows: [ 150 x 4 | 1 ] #> PC1 PC2 PC3 ... | species #> | <fct> #> 1 -2.26 -0.478 0.127 | 1 setosa #> 2 -2.07 0.672 0.234 ... | 2 setosa #> 3 -2.36 0.341 -0.0441 | 3 setosa #> 4 -2.29 0.595 -0.0910 | 4 setosa #> 5 -2.38 -0.645 -0.0157 | 5 setosa #> # … with 145 more rows #> # #> # Columns: [ 4 x 4 | 0 ] #> PC1 PC2 PC3 ... | #> | #> 1 0.521 -0.377 0.720 | #> 2 -0.269 -0.923 -0.244 ... | #> 3 0.580 -0.0245 -0.142 | #> 4 0.565 -0.0669 -0.634 |# row-principal biplot with centroids and confidence ellipses iris_pca %>% ggbiplot(aes(color = species)) + theme_bw() + scale_color_brewer(type = "qual", palette = 2) + geom_rows_point(alpha = .5) + stat_rows_center(fun.center = "mean", size = 3, shape = "triangle") + stat_rows_ellipse(level = .99) + ggtitle( "Row-principal PCA biplot of Anderson iris measurements", "Overlaid with centroids and 99% confidence ellipses" )# row-principal biplot with centroids and confidence elliptical disks iris_pca %>% ggbiplot(aes(color = species)) + theme_bw() + geom_rows_point() + geom_polygon( aes(fill = species), color = NA, alpha = .25, stat = "rows_ellipse" ) + geom_cols_vector(color = "#444444") + scale_color_brewer( type = "qual", palette = 2, aesthetics = c("color", "fill") ) + ggtitle( "Row-principal PCA biplot of Anderson iris measurements", "Overlaid with 95% confidence disks" )